Stephan Michael Feller
Prof. Stephan Michael Feller
Institut für Molekulare Medizin
Sektion Tumorbiologie
ZAMED (Zentrum für Angewandte Medizinische
und Humanbiologische Forschung)
Heinrich-Damerow-Str. 1
06120 Halle (Saale)
phone: +49 (0) 345-55 22915
stephan.feller@medizin.uni-halle.de
Analysis of interactions, subcellular localizations and translocations of signal-computing protein complexes in normal cells and cancers
Our project investigates the molecular mechanisms of signal processing in cells as we have a only marginal understanding of the higher order signal integration processes in cells so far. Grasping these molecular computations in much more detail may ultimately lead to new strategies for inhibiting pathological signaling processes in cancers and other human diseases.
Brief introduction
Cells are composed of numerous distinct compartments separated in most cases by membrane structures. This compartimentalization allows the re-use of proteins in several distinct biological contexts as well as creating specific environments with respect to pH, redox status, protein and lipid composition etc.
The specific functions of cellular proteins in different subcellular compartments remain poorly studied to date. Analysing distinct pools of proteins, their modification status changes, their interaction partners and their translocation can therefore provide essential clues about physiological and pathological regulatory processes at the ultrastructural/molecular and functional levels.
A major focus of our research team is on Gab family proteins. These function as platform proteins in the formation of large protein complexes downstream of oncogenic receptor tyrosine kinases like those of the c-Met and EGFR/ErbB families but are also essential for a range of physiological processes during embryonic and fetal development.
Research project
We have recently proposed an entirely novel, well-received model of how signal computation is enabled by protein complexes assembled on Gab proteins (Simister et al. 2011, PLOS Biol 9:e1000591, Lewitzky et al. 2012, FEBS Lett 586, 2740; see also Figure 1).
We will now apply a range of biochemcial, biophysical and cell biology methods to further investigate the dynamic molecular actions and interactions of the Gab-mediated protein complexes and their shuttling between different subcellular locations (project collaborators include Prof. Fred Schaper, OVGU Magdeburg and Prof. Stefan Knapp, Oxford University).
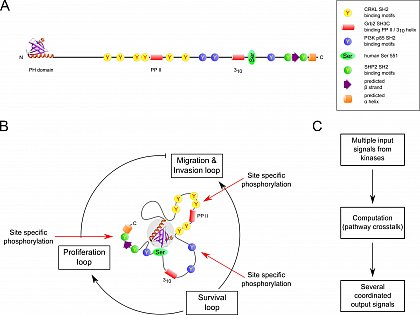
Figure 1
Human Gab1 protein in the traditional stick representation and as a graphic display of its presumed signal computation architecture according to the N-terminal folding nucleation (NFN) hypothesis.
(A) ‘Classical’ stick representation of human Gab1. (B) Schematic representation of the proposed Gab1 signal computation architecture. (C) Simple functional diagram of the presumed signal processing via the Gab1 LMD protein platform-based multi-protein complex.
For a full explanation of all details, please refer to Lewitzky et al. 2012, FEBS Lett 586, 2740.
